建立超图 怎样建立一个超图详解
ran_梦 人气:01.图和超图
图作为一种数据结构,由节点和边组成,可由下图表示。其中一个边只能链接两个节点。一个图可表示为G=(v,e,w)
其中v表示节点,e表示边,w表示节点的特征。关于图的表示可参考,本文不再详述。
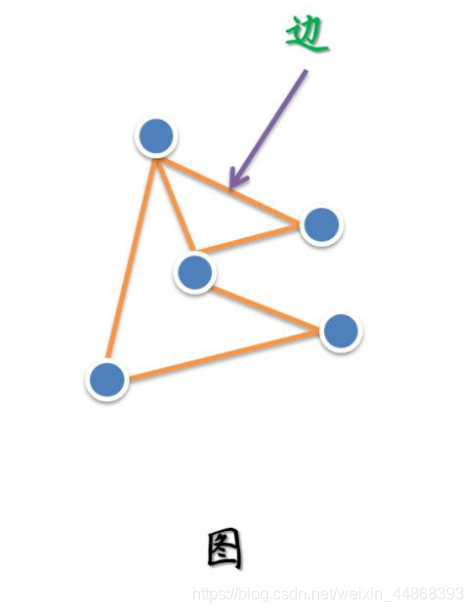
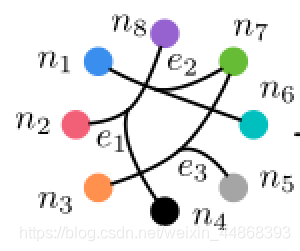
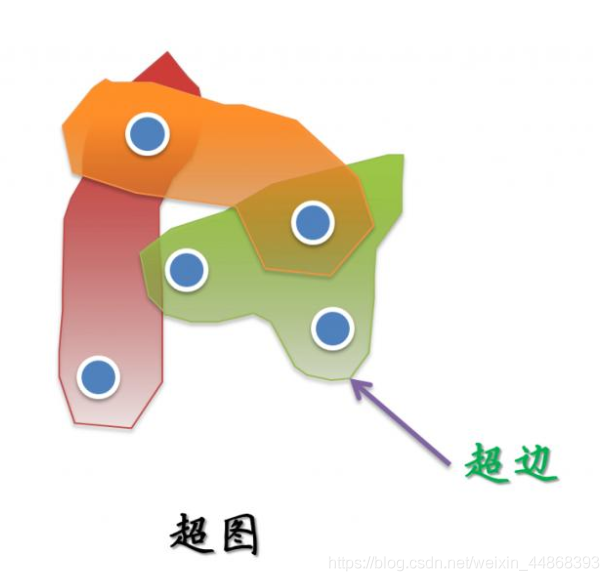
对于超图,其与图结构最主要的区别就是一条边可以连接多个节点,因此我们可以认为图是一种特殊的超图。超图结构如下图所示。
超图可表示为G=(υ,ε,ω)。其中υ为节点集合,ε为超边集合,ω为超边权重的对称矩阵。超图G可以关联矩阵H来表示,其词条定义为:
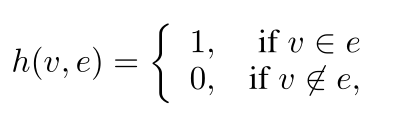
改公式可解释为如果某个节点属于某个超边,则关联矩阵H的值为1,否则为0。
对于单个节点v可定义为:
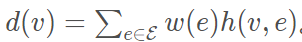
可解释为连接该节点的所有边乘上权重向量的和。
Dₑ和Dᵥ由d(v)和s(e)分别表示为超边和节点的对角矩阵。
单个边可定义为:
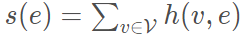
可以理解为该边包含的所有节点之和。
2.实例
下面举出一个具体实例帮助理解超图的构建。以该图为例

图中有8个节点,3个超边。超边的细化图如下:
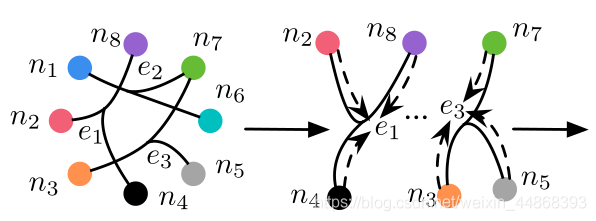
假设权重&W&为全1矩阵,因为它对构建超图数据结果无影响,那么H为一个3行8列的矩阵,表示为:
h(1,1) = 0
h(2,1) = 1
h(3,1) = 0
h(4,1) = 1
h(5,1) = 0
h(6,1) = 0
h(7,1) = 0
h(8,1) = 1
h(1,2) = 1
h(2,2) = 0
h(3,2) = 0
h(4,2) = 0
h(5,2) = 0
h(6,2) = 1
h(7,2) = 1
h(8,2) = 0
h(1,3) = 0
h(2,3) = 0
h(3,3) = 1
h(4,3) = 0
h(5,3) = 1
h(6,3) = 0
h(7,3) = 1
h(8,3) = 0
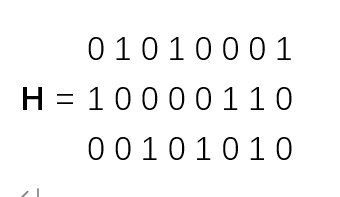
De表示为:
d(1) = 1
d(2) = 1
d(3) = 1
d(4) = 1
d(5) = 1
d(6) = 1
d(7) = 2
d(8) = 1
Dv表示为:
s(1) = 3
s(2) = 3
s(3) = 3
3.代码实现
下面我们用python代码进行编程,我们的目标是在知道节点的特征W通过特征的距离来生成 G \mathcal{G} G矩阵。路线为:W,H, G \mathcal{G} G。主要代码如下:
import numpy as np
#KNN生成H
x = np.array([[1,0,0,0,1,0,1,0,0,0],
[1,1,1,0,0,0,1,1,1,0],
[1,1,1,0,0,1,1,1,1,0],
[0,1,0,0,0,0,1,0,1,0],
[1,1,1,1,0,0,1,1,0,1],
[1,0,1,0,0,1,0,1,1,0],
[0,1,0,0,1,0,1,1,1,0],
[0,1,1,0,1,0,1,0,1,1]])
def Eu_dis(x):
"""
Calculate the distance among each raw of x
:param x: N X D
N: the object number
D: Dimension of the feature
:return: N X N distance matrix
"""
x = np.mat(x)
aa = np.sum(np.multiply(x, x), 1)
ab = x * x.T
dist_mat = aa + aa.T - 2 * ab
dist_mat[dist_mat < 0] = 0
dist_mat = np.sqrt(dist_mat)
dist_mat = np.maximum(dist_mat, dist_mat.T)
return dist_mat
def hyperedge_concat(*H_list):
"""
Concatenate hyperedge group in H_list
:param H_list: Hyperedge groups which contain two or more hypergraph incidence matrix
:return: Fused hypergraph incidence matrix
"""
H = None
for h in H_list:
if h is not None and h != []:
# for the first H appended to fused hypergraph incidence matrix
if H is None:
H = h
else:
if type(h) != list:
H = np.hstack((H, h))
else:
tmp = []
for a, b in zip(H, h):
tmp.append(np.hstack((a, b)))
H = tmp
return H
def construct_H_with_KNN_from_distance(dis_mat, k_neig, is_probH=True, m_prob=1):
"""
construct hypregraph incidence matrix from hypergraph node distance matrix
:param dis_mat: node distance matrix
:param k_neig: K nearest neighbor
:param is_probH: prob Vertex-Edge matrix or binary
:param m_prob: prob
:return: N_object X N_hyperedge
"""
n_obj = dis_mat.shape[0]
# construct hyperedge from the central feature space of each node
n_edge = n_obj
H = np.zeros((n_obj, n_edge))
for center_idx in range(n_obj):
dis_mat[center_idx, center_idx] = 0
dis_vec = dis_mat[center_idx]
nearest_idx = np.array(np.argsort(dis_vec)).squeeze()
avg_dis = np.average(dis_vec)
if not np.any(nearest_idx[:k_neig] == center_idx):
nearest_idx[k_neig - 1] = center_idx
for node_idx in nearest_idx[:k_neig]:
if is_probH:
H[node_idx, center_idx] = np.exp(-dis_vec[0, node_idx] ** 2 / (m_prob * avg_dis) ** 2)
else:
H[node_idx, center_idx] = 1.0
return H
def construct_H_with_KNN(X, K_neigs=[10], split_diff_scale=False, is_probH=True, m_prob=1):
"""
init multi-scale hypergraph Vertex-Edge matrix from original node feature matrix
:param X: N_object x feature_number
:param K_neigs: the number of neighbor expansion
:param split_diff_scale: whether split hyperedge group at different neighbor scale
:param is_probH: prob Vertex-Edge matrix or binary
:param m_prob: prob
:return: N_object x N_hyperedge
"""
if len(X.shape) != 2:
X = X.reshape(-1, X.shape[-1])
if type(K_neigs) == int:
K_neigs = [K_neigs]
dis_mat = Eu_dis(X)
H = []
for k_neig in K_neigs:
H_tmp = construct_H_with_KNN_from_distance(dis_mat, k_neig, is_probH, m_prob)
if not split_diff_scale:
H = hyperedge_concat(H, H_tmp)
else:
H.append(H_tmp)
return H
H = construct_H_with_KNN(x)
#生成G
def generate_G_from_H(H, variable_weight=False):
"""
calculate G from hypgraph incidence matrix H
:param H: hypergraph incidence matrix H
:param variable_weight: whether the weight of hyperedge is variable
:return: G
"""
if type(H) != list:
return _generate_G_from_H(H, variable_weight)
else:
G = []
for sub_H in H:
G.append(generate_G_from_H(sub_H, variable_weight))
return G
def _generate_G_from_H(H, variable_weight=False):
"""
calculate G from hypgraph incidence matrix H
:param H: hypergraph incidence matrix H
:param variable_weight: whether the weight of hyperedge is variable
:return: G
"""
H = np.array(H)
n_edge = H.shape[1]
# the weight of the hyperedge
W = np.ones(n_edge)
# the degree of the node
DV = np.sum(H * W, axis=1)
# the degree of the hyperedge
DE = np.sum(H, axis=0)
invDE = np.mat(np.diag(np.power(DE, -1)))
DV2 = np.mat(np.diag(np.power(DV, -0.5)))
W = np.mat(np.diag(W))
H = np.mat(H)
HT = H.T
if variable_weight:
DV2_H = DV2 * H
invDE_HT_DV2 = invDE * HT * DV2
return DV2_H, W, invDE_HT_DV2
else:
G = DV2 * H * W * invDE * HT * DV2
return G
G = generate_G_from_H(H)
实验结果:
H

G

加载全部内容