Python3实现打格点算法 Python3实现打格点算法的GPU加速实例详解
DECHIN 人气:0技术背景
在数学和物理学领域,总是充满了各种连续的函数模型。而当我们用现代计算机的技术去处理这些问题的时候,事实上是无法直接处理连续模型的,绝大多数的情况下都要转化成一个离散的模型再进行数值的计算。比如计算数值的积分,计算数值的二阶导数(海森矩阵)等等。这里我们所介绍的打格点的算法,正是一种典型的离散化方法。这个对空间做离散化的方法,可以在很大程度上简化运算量。比如在分子动力学模拟中,计算近邻表的时候,如果不采用打格点的方法,那么就要针对整个空间所有的原子进行搜索,计算出来距离再判断是否近邻。而如果采用打格点的方法,我们只需要先遍历一遍原子对齐进行打格点的离散化,之后再计算近邻表的时候,只需要计算三维空间下邻近的27个格子中的原子是否满足近邻条件即可。在这篇文章中,我们主要探讨如何用GPU来实现打格点的算法。
打格点算法实现
我们先来用一个例子说明一下什么叫打格点。对于一个给定所有原子坐标的系统,也就是已知了[x,y,z],我们需要得到的是这些原子所在的对应的格子位置[nx,ny,nz]。我们先看一下在CPU上的实现方案,是一个遍历一次的算法:
# cuda_grid.py
from numba import jit
from numba import cuda
import numpy as np
def grid_by_cpu(crd, rxyz, atoms, grids):
"""Transform coordinates [x,y,z] into grids [nx,ny,nz].
Args:
crd(list): The 3-D coordinates of atoms.
rxyz(list): The list includes xmin,ymin,zmin,grid_num.
atoms(int): The total number of atoms.
grids(list): The transformed grids matrix.
"""
for i in range(atoms):
grids[i][0] = int((crd[i][0]-rxyz[0])/rxyz[3])
grids[i][1] = int((crd[i][1]-rxyz[1])/rxyz[3])
grids[i][2] = int((crd[i][2]-rxyz[2])/rxyz[3])
return grids
if __name__=='__main__':
np.random.seed(1)
atoms = 4
grid_size = 0.1
crd = np.random.random((atoms,3)).astype(np.float32)
xmin = min(crd[:,0])
ymin = min(crd[:,1])
zmin = min(crd[:,2])
xmax = max(crd[:,0])
ymax = max(crd[:,1])
zmax = max(crd[:,2])
xgrids = int((xmax-xmin)/grid_size)+1
ygrids = int((ymax-ymin)/grid_size)+1
zgrids = int((zmax-zmin)/grid_size)+1
rxyz = np.array([xmin,ymin,zmin,grid_size], dtype=np.float32)
grids = np.ones_like(crd)*(-1)
grids = grids.astype(np.float32)
grids_cpu = grid_by_cpu(crd, rxyz, atoms, grids)
print (crd)
print (grids_cpu)
import matplotlib.pyplot as plt
plt.figure()
plt.plot(crd[:,0], crd[:,1], 'o', color='red')
for grid in range(ygrids+1):
plt.plot([xmin,xmin+grid_size*xgrids], [ymin+grid_size*grid,ymin+grid_size*grid], color='black')
for grid in range(xgrids+1):
plt.plot([xmin+grid_size*grid,xmin+grid_size*grid], [ymin,ymin+grid_size*ygrids], color='black')
plt.savefig('Atom_Grids.png')
输出结果如下,
$ python3 cuda_grid.py
[[4.17021990e-01 7.20324516e-01 1.14374816e-04]
[3.02332580e-01 1.46755889e-01 9.23385918e-02]
[1.86260208e-01 3.45560730e-01 3.96767467e-01]
[5.38816750e-01 4.19194520e-01 6.85219526e-01]]
[[2. 5. 0.]
[1. 0. 0.]
[0. 1. 3.]
[3. 2. 6.]]
上面两个打印输出就分别对应于[x,y,z]和[nx,ny,nz],比如第一个原子被放到了编号为[2,5,0]的格点。那么为了方便理解打格点的方法,我们把这个三维空间的原子系统和打格点以后的标号取前两个维度来可视化一下结果,作图以后效果如下:
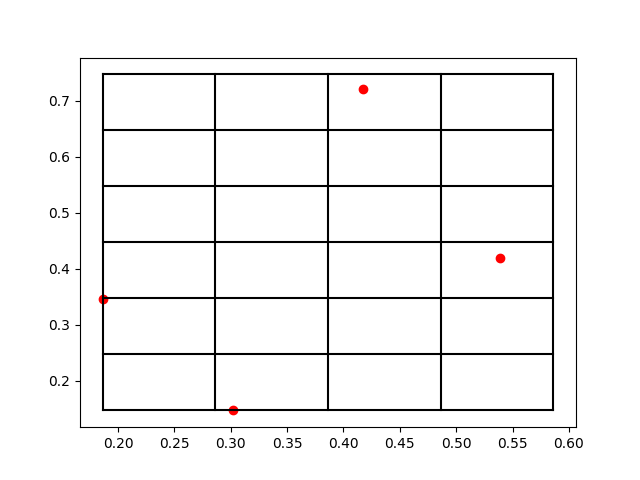
我们可以看到,这些红色的点就是原子所处的位置,而黑色的网格线就是我们所标记的格点。在原子数量比较多的时候,有可能出现在一个网格中存在很多个原子的情况,所以如何打格点,格点大小如何去定义,这都是不同场景下的经验参数,需要大家一起去摸索。
打格点算法加速
在上面这个算法实现中,我们主要是用到了一个for循环,这时候我们可以想到numba所支持的向量化运算,还有GPU硬件加速,这里我们先对比一下三种实现方案的计算结果:
# cuda_grid.py
from numba import jit
from numba import cuda
import numpy as np
def grid_by_cpu(crd, rxyz, atoms, grids):
"""Transform coordinates [x,y,z] into grids [nx,ny,nz].
Args:
crd(list): The 3-D coordinates of atoms.
rxyz(list): The list includes xmin,ymin,zmin,grid_num.
atoms(int): The total number of atoms.
grids(list): The transformed grids matrix.
"""
for i in range(atoms):
grids[i][0] = int((crd[i][0]-rxyz[0])/rxyz[3])
grids[i][1] = int((crd[i][1]-rxyz[1])/rxyz[3])
grids[i][2] = int((crd[i][2]-rxyz[2])/rxyz[3])
return grids
@jit
def grid_by_jit(crd, rxyz, atoms, grids):
"""Transform coordinates [x,y,z] into grids [nx,ny,nz].
Args:
crd(list): The 3-D coordinates of atoms.
rxyz(list): The list includes xmin,ymin,zmin,grid_num.
atoms(int): The total number of atoms.
grids(list): The transformed grids matrix.
"""
for i in range(atoms):
grids[i][0] = int((crd[i][0]-rxyz[0])/rxyz[3])
grids[i][1] = int((crd[i][1]-rxyz[1])/rxyz[3])
grids[i][2] = int((crd[i][2]-rxyz[2])/rxyz[3])
return grids
@cuda.jit
def grid_by_gpu(crd, rxyz, grids):
"""Transform coordinates [x,y,z] into grids [nx,ny,nz].
Args:
crd(list): The 3-D coordinates of atoms.
rxyz(list): The list includes xmin,ymin,zmin,grid_num.
atoms(int): The total number of atoms.
grids(list): The transformed grids matrix.
"""
i,j = cuda.grid(2)
grids[i][j] = int((crd[i][j]-rxyz[j])/rxyz[3])
if __name__=='__main__':
np.random.seed(1)
atoms = 4
grid_size = 0.1
crd = np.random.random((atoms,3)).astype(np.float32)
xmin = min(crd[:,0])
ymin = min(crd[:,1])
zmin = min(crd[:,2])
xmax = max(crd[:,0])
ymax = max(crd[:,1])
zmax = max(crd[:,2])
xgrids = int((xmax-xmin)/grid_size)+1
ygrids = int((ymax-ymin)/grid_size)+1
zgrids = int((zmax-zmin)/grid_size)+1
rxyz = np.array([xmin,ymin,zmin,grid_size], dtype=np.float32)
crd_cuda = cuda.to_device(crd)
rxyz_cuda = cuda.to_device(rxyz)
grids = np.ones_like(crd)*(-1)
grids = grids.astype(np.float32)
grids_cpu = grid_by_cpu(crd, rxyz, atoms, grids)
grids = np.ones_like(crd)*(-1)
grids_jit = grid_by_jit(crd, rxyz, atoms, grids)
grids = np.ones_like(crd)*(-1)
grids_cuda = cuda.to_device(grids)
grid_by_gpu[(atoms,3),(1,1)](crd_cuda,
rxyz_cuda,
grids_cuda)
print (crd)
print (grids_cpu)
print (grids_jit)
print (grids_cuda.copy_to_host())
输出结果如下:
$ python3 cuda_grid.py
/home/dechin/anaconda3/lib/python3.8/site-packages/numba/cuda/compiler.py:865: NumbaPerformanceWarning: Grid size (12) < 2 * SM count (72) will likely result in GPU under utilization due to low occupancy.
warn(NumbaPerformanceWarning(msg))
[[4.17021990e-01 7.20324516e-01 1.14374816e-04]
[3.02332580e-01 1.46755889e-01 9.23385918e-02]
[1.86260208e-01 3.45560730e-01 3.96767467e-01]
[5.38816750e-01 4.19194520e-01 6.85219526e-01]]
[[2. 5. 0.]
[1. 0. 0.]
[0. 1. 3.]
[3. 2. 6.]]
[[2. 5. 0.]
[1. 0. 0.]
[0. 1. 3.]
[3. 2. 6.]]
[[2. 5. 0.]
[1. 0. 0.]
[0. 1. 3.]
[3. 2. 6.]]
我们先看到这里面的告警信息,因为GPU硬件加速要在一定密度的运算量之上才能够有比较明显的加速效果。比如说我们只是计算两个数字的加和,那么是完全没有必要使用到GPU的。但是如果我们要计算两个非常大的数组的加和,那么这个时候GPU就能够发挥出非常大的价值。因为这里我们的案例中只有4个原子,因此提示我们这时候是体现不出来GPU的加速效果的。我们仅仅关注下这里的运算结果,在不同体系下得到的格点结果是一致的,那么接下来就可以对比一下几种不同实现方式的速度差异。
# cuda_grid.py
from numba import jit
from numba import cuda
import numpy as np
def grid_by_cpu(crd, rxyz, atoms, grids):
"""Transform coordinates [x,y,z] into grids [nx,ny,nz].
Args:
crd(list): The 3-D coordinates of atoms.
rxyz(list): The list includes xmin,ymin,zmin,grid_num.
atoms(int): The total number of atoms.
grids(list): The transformed grids matrix.
"""
for i in range(atoms):
grids[i][0] = int((crd[i][0]-rxyz[0])/rxyz[3])
grids[i][1] = int((crd[i][1]-rxyz[1])/rxyz[3])
grids[i][2] = int((crd[i][2]-rxyz[2])/rxyz[3])
return grids
@jit
def grid_by_jit(crd, rxyz, atoms, grids):
"""Transform coordinates [x,y,z] into grids [nx,ny,nz].
Args:
crd(list): The 3-D coordinates of atoms.
rxyz(list): The list includes xmin,ymin,zmin,grid_num.
atoms(int): The total number of atoms.
grids(list): The transformed grids matrix.
"""
for i in range(atoms):
grids[i][0] = int((crd[i][0]-rxyz[0])/rxyz[3])
grids[i][1] = int((crd[i][1]-rxyz[1])/rxyz[3])
grids[i][2] = int((crd[i][2]-rxyz[2])/rxyz[3])
return grids
@cuda.jit
def grid_by_gpu(crd, rxyz, grids):
"""Transform coordinates [x,y,z] into grids [nx,ny,nz].
Args:
crd(list): The 3-D coordinates of atoms.
rxyz(list): The list includes xmin,ymin,zmin,grid_num.
atoms(int): The total number of atoms.
grids(list): The transformed grids matrix.
"""
i,j = cuda.grid(2)
grids[i][j] = int((crd[i][j]-rxyz[j])/rxyz[3])
if __name__=='__main__':
import time
from tqdm import trange
np.random.seed(1)
atoms = 100000
grid_size = 0.1
crd = np.random.random((atoms,3)).astype(np.float32)
xmin = min(crd[:,0])
ymin = min(crd[:,1])
zmin = min(crd[:,2])
xmax = max(crd[:,0])
ymax = max(crd[:,1])
zmax = max(crd[:,2])
xgrids = int((xmax-xmin)/grid_size)+1
ygrids = int((ymax-ymin)/grid_size)+1
zgrids = int((zmax-zmin)/grid_size)+1
rxyz = np.array([xmin,ymin,zmin,grid_size], dtype=np.float32)
crd_cuda = cuda.to_device(crd)
rxyz_cuda = cuda.to_device(rxyz)
cpu_time = 0
jit_time = 0
gpu_time = 0
for i in trange(100):
grids = np.ones_like(crd)*(-1)
grids = grids.astype(np.float32)
time0 = time.time()
grids_cpu = grid_by_cpu(crd, rxyz, atoms, grids)
time1 = time.time()
grids = np.ones_like(crd)*(-1)
time2 = time.time()
grids_jit = grid_by_jit(crd, rxyz, atoms, grids)
time3 = time.time()
grids = np.ones_like(crd)*(-1)
grids_cuda = cuda.to_device(grids)
time4 = time.time()
grid_by_gpu[(atoms,3),(1,1)](crd_cuda,
rxyz_cuda,
grids_cuda)
time5 = time.time()
if i != 0:
cpu_time += time1 - time0
jit_time += time3 - time2
gpu_time += time5 - time4
print ('The time cost of CPU calculation is: {}s'.format(cpu_time))
print ('The time cost of JIT calculation is: {}s'.format(jit_time))
print ('The time cost of GPU calculation is: {}s'.format(gpu_time))
输出结果如下:
$ python3 cuda_grid.py
100%|███████████████████████████| 100/100 [00:23<00:00, 4.18it/s]
The time cost of CPU calculation is: 23.01943016052246s
The time cost of JIT calculation is: 0.04810166358947754s
The time cost of GPU calculation is: 0.01806473731994629s
在100000个原子的体系规模下,普通的for循环实现效率就非常的低下,需要23s,而经过向量化运算的加速之后,直接飞升到了0.048s,而GPU上的加速更是达到了0.018s,相比于没有GPU硬件加速的场景,实现了将近2倍的加速。但是这还远远不是GPU加速的上限,让我们再测试一个更大的案例:
# cuda_grid.py
from numba import jit
from numba import cuda
import numpy as np
def grid_by_cpu(crd, rxyz, atoms, grids):
"""Transform coordinates [x,y,z] into grids [nx,ny,nz].
Args:
crd(list): The 3-D coordinates of atoms.
rxyz(list): The list includes xmin,ymin,zmin,grid_num.
atoms(int): The total number of atoms.
grids(list): The transformed grids matrix.
"""
for i in range(atoms):
grids[i][0] = int((crd[i][0]-rxyz[0])/rxyz[3])
grids[i][1] = int((crd[i][1]-rxyz[1])/rxyz[3])
grids[i][2] = int((crd[i][2]-rxyz[2])/rxyz[3])
return grids
@jit
def grid_by_jit(crd, rxyz, atoms, grids):
"""Transform coordinates [x,y,z] into grids [nx,ny,nz].
Args:
crd(list): The 3-D coordinates of atoms.
rxyz(list): The list includes xmin,ymin,zmin,grid_num.
atoms(int): The total number of atoms.
grids(list): The transformed grids matrix.
"""
for i in range(atoms):
grids[i][0] = int((crd[i][0]-rxyz[0])/rxyz[3])
grids[i][1] = int((crd[i][1]-rxyz[1])/rxyz[3])
grids[i][2] = int((crd[i][2]-rxyz[2])/rxyz[3])
return grids
@cuda.jit
def grid_by_gpu(crd, rxyz, grids):
"""Transform coordinates [x,y,z] into grids [nx,ny,nz].
Args:
crd(list): The 3-D coordinates of atoms.
rxyz(list): The list includes xmin,ymin,zmin,grid_num.
atoms(int): The total number of atoms.
grids(list): The transformed grids matrix.
"""
i,j = cuda.grid(2)
grids[i][j] = int((crd[i][j]-rxyz[j])/rxyz[3])
if __name__=='__main__':
import time
from tqdm import trange
np.random.seed(1)
atoms = 5000000
grid_size = 0.1
crd = np.random.random((atoms,3)).astype(np.float32)
xmin = min(crd[:,0])
ymin = min(crd[:,1])
zmin = min(crd[:,2])
xmax = max(crd[:,0])
ymax = max(crd[:,1])
zmax = max(crd[:,2])
xgrids = int((xmax-xmin)/grid_size)+1
ygrids = int((ymax-ymin)/grid_size)+1
zgrids = int((zmax-zmin)/grid_size)+1
rxyz = np.array([xmin,ymin,zmin,grid_size], dtype=np.float32)
crd_cuda = cuda.to_device(crd)
rxyz_cuda = cuda.to_device(rxyz)
jit_time = 0
gpu_time = 0
for i in trange(100):
grids = np.ones_like(crd)*(-1)
time2 = time.time()
grids_jit = grid_by_jit(crd, rxyz, atoms, grids)
time3 = time.time()
grids = np.ones_like(crd)*(-1)
grids_cuda = cuda.to_device(grids)
time4 = time.time()
grid_by_gpu[(atoms,3),(1,1)](crd_cuda,
rxyz_cuda,
grids_cuda)
time5 = time.time()
if i != 0:
jit_time += time3 - time2
gpu_time += time5 - time4
print ('The time cost of JIT calculation is: {}s'.format(jit_time))
print ('The time cost of GPU calculation is: {}s'.format(gpu_time))
在这个5000000个原子的案例中,因为普通的for循环已经实在是跑不动了,因此我们就干脆不统计这一部分的时间,最后输出结果如下:
$ python3 cuda_grid.py
100%|███████████████████████████| 100/100 [00:09<00:00, 10.15it/s]
The time cost of JIT calculation is: 2.3743042945861816s
The time cost of GPU calculation is: 0.022843599319458008s
在如此大规模的运算下,GPU实现100倍的加速,而此时作为对比的CPU上的实现方法是已经用上了向量化运算的操作,也已经可以认为是一个极致的加速了。
总结概要
在这篇文章中,我们主要介绍了打格点算法在分子动力学模拟中的重要价值,以及几种不同的实现方式。其中最普通的for循环的实现效率比较低下,从算法复杂度上来讲却已经是极致。而基于CPU上的向量化运算的技术,可以对计算过程进行非常深度的优化。当然,这个案例在不同的硬件上也能够发挥出明显不同的加速效果,在GPU的加持之下,可以获得100倍以上的加速效果。这也是一个在Python上实现GPU加速算法的一个典型案例。
加载全部内容